Who's your daddy? Paternal Mitochondria
The Scientist reports that paternal mitochondria have been found in the muscle cells of a sufferer from myopathy, by a team from the Beth Israel Deaconess Medical Center and Harvard Medical School. This means that paternal mitochondria not only find their way into the oocyte (the female egg cell), as Ankel-Simons and Cummins reported in 1996, but also can continue into the mature organism. What effect does it have on Mitochondrial Eve?
The article goes on to say
Investigating the basis for a mitchondrial myopathy in a patient described in 2002, members of Kraytsberg’s group had discovered that his muscle contained about 90% paternal mtDNA carrying a detrimental mutation responsible for the disease, Khrapko told The Scientist by email.
The abbreviation “mt” is biologist speak for “mitochondrial”.
Mitochondrial Eve is the popular name given for the last female common ancestor of all humans. Since (it was assumed) mtDNA (the DNA contained in the mitochondria themselves) was only inherited by the mother’s lineage, we could work out how long since that person lived, as a way of clocking human history. Because it was asexually transmitted - which is to say it didn’t recombine paternal and maternal mtDNA, and assuming that the mutation rate was roughly constant, the theory was able to estimate the age of that population at 200,000 years before present.
Except, it turns out, not only does paternal mtDNA find its way into progeny, it even recombines with maternal mtDNA, muddying the “signal”.
The “Y chromosome Adam” lived only 35,000 to 90,000 years ago. This is more in keeping with what we know of the date of modern humans (around 120,000 years ago), although there is no reason why mt genes could not persist over a speciation event.
I asked Reed Cartright - a real biologist - to work out what effect paternal recombination of mtDNA means to Mitochondrial Eve. It turns out, as mathematical modeling often does, to be surprising. It makes very little difference at all. The following graph shows that until the rate of individual recombination is very high, paternal mtDNA does very little to estimates of mtEve.
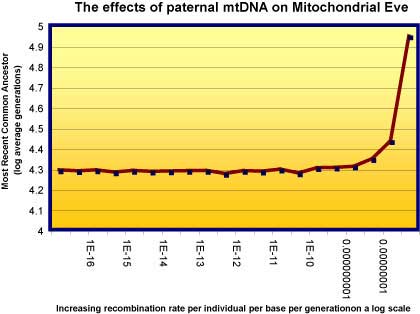
The graph shows how the recombination of nuclear DNA affects estimates of the most recent common ancestor (TMRCA). 1e-8 (1 in 10 to the 8th power, or one in each 100 million) is the estimate for nuclear recombination rates. MTDNA rates are for many reasons many-fold less than this. So the effect is at worst minimal, and at best affects mtEve not at all.
(See Reed’s [URL = http://www.pandasthumb.org/pt-archives/000234.html#more]EvoMath 3: Genetic Drift and Coalescence, Briefly[/URL] entry for a detaile discussion of the phenomenon of “coalescence”, where genes drift into fixation replacing other genes, which is what is occurring with mtEve’s mitochondrial DNA.)
Anyway, it’s an interesting development. In recent years, the often-asserted textbook idea that paternal mtDNA is never transmitted to progeny has been shown to be incorrect in a number of species, and this has been known for many years now. The blue mussel, Mytilus edulis has maternal mtDNA in the body tissue of males, but paternal mtDNA in their gonads, and the sperm mitochondria are randomly dispersed in female progeny (Cao et al. 2004). The parasite that causes schistosomiasis inherits biparental mtDNA, which is not eliminated if the progeny are species hybrids (Jannotti-Pannos et al. 2001). And paternal inheritance has been identified in the great tit, Parus major (Kvist et al. 2003), another mussel Mytilus galloprovincialis (Ladoukakis et al., 2002) among others both sea mussell and fresh water mussells(Zouros 2000).
Nevertheless, the evidence is that mtDNA is eliminated in most species of vertebrates, partly because paternal mitochondria are “tagged” in the oocyte (maternal egg) after fertilisation (Shitara, 2000; Sutovsky, 2000; Sutovsky, 2003; Sutovsky, et al, 2003; Sutovsky et al., 2004; Cummins, 2000). Paternal mitochondrial transmission appears to inhibit mammalian spermatogenesis, and patern inheritance appears, as in the case we started with, to cause diseases, including muscular myopathy (Johns, 2003; Gianotten, 2003). There is evidence that mitochondria in males suffer greater mutation rates. Because mitochondria do not have the repair mechanisms of the nuclear genetic machinery, uniparental inheritance may be due to selection against biparental transmission (Hoekstra, 2000).
So the topic turns out to be evolutionarily relevant in another way, and reaffirms the old rule in biology: there’s nothing so absurd or unusual that it doesn’t exist in at least one organism. Rules in biology, like “Paternal mtDNA is not inherited” are not laws of nature, they are rules of thumb. We learn when we learn the exceptions, rather than “falsify” what we already knew.
References
Ankel-Simons, F. and J. M. Cummins (1996). “Misconceptions about mitochondria and mammalian fertilization: Implications for theories on human evolution.” Proc. Natl. Acad. Sci. USA 93(November): 13859-13863.
Cao, L., E. Kenchington, et al. (2004). “Differential segregation patterns of sperm mitochondria in embryos of the blue mussel (Mytilus edulis).” Genetics 166(2): 883-94.
Cummins, J. M. (2000). “Fertilization and elimination of the paternal mitochondrial genome.” Hum Reprod 15 Suppl 2: 92-101.
Gianotten, J., M. J. Hoffer, et al. (2003). “Partial DAZ deletions in a family with five infertile brothers.” Fertil Steril 79 Suppl 3: 1652-5.
Hoekstra, R. F. (2000). “Evolutionary origin and consequences of uniparental mitochondrial inheritance.” Hum Reprod 15 Suppl 2: 102-11.
Jannotti-Passos, L. K., C. P. Souza, et al. (2001). “Biparental mitochondrial DNA inheritance in the parasitic trematode Schistosoma mansoni.” J Parasitol 87(1): 79-82.
Johns, D. R. (2003). “Paternal transmission of mitochondrial DNA is (fortunately) rare.” Ann Neurol 54(4): 422-4.
Kvist, L., J. Martens, et al. (2003). “Paternal leakage of mitochondrial DNA in the great tit (Parus major).” Mol Biol Evol 20(2): 243-7.
Ladoukakis, E. D., C. Saavedra, et al. (2002). “Mitochondrial DNA variation in a species with two mitochondrial genomes: the case of Mytilus galloprovincialis from the Atlantic, the Mediterranean and the Black Sea.” Mol Ecol 11(4): 755-69.
Shitara, H., H. Kaneda, et al. (2000). “Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis.” Genetics 156(3): 1277-84.
Sutovsky, P. (2003). “Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: killing three birds with one stone.” Microsc Res Tech 61(1): 88-102.
Sutovsky, P., K. Van Leyen, et al. (2004). “Degradation of paternal mitochondria after fertilization: implications for heteroplasmy, assisted reproductive technologies and mtDNA inheritance.” Reprod Biomed Online 8(1): 24-33.
Sutovsky, P., R. D. Moreno, et al. (2000). “Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos.” Biol Reprod 63(2): 582-90.
Sutovsky, P., T. C. McCauley, et al. (2003). “Early degradation of paternal mitochondria in domestic pig (Sus scrofa) is prevented by selective proteasomal inhibitors lactacystin and MG132.” Biol Reprod 68(5): 1793-800.
Zouros, E. (2000). “The exceptional mitochondrial DNA system of the mussel family Mytilidae.” Genes Genet Syst 75(6): 313-8.
Thanks to Reed for his help in getting a liberal arts major to understand this…
