A study in Science has returned biological methods to linguistic evolution in a reversal of history, and concluded that one can, within limits, reconstruct the history of language.
Charles Darwin was not the first person to suppose that historical evolution could be recognised by homologies and represented by tree diagrams. That honour goes to Sir William Jones in 1797, although the tree idea was later.
Jones argued that one could compare cognate terms and infer a historical relationship between languages and this has become the foundation of modern philology. For example, words that are based on the idea of "knowing" (including, as it happens, "idea") generate a tree of Indo-European languages. [And like biological evolution, there are "creationists" who think that all language was created in Sanskrit.]
Now, a study in Science has returned biological methods to linguistic evolution in a reversal of history, and come up with some interesting conclusions.
This paper is not the first to do this, of course. Merritt Ruhlen previously argued that all human languages could be traced back to a last common ancestral tongue, presumably the original language in Africa. However, many linguists think he not only chose cognate terms too broadly to bolster his reconstruction, but he also overlooked a singular problem for all historical sciences: time erases information. Still, the idea of constructing a phylogenetic tree for language is feasible, to some depth. What that depth is, and the rate at which words evolve, is the subject of the new paper.
The current study [available here but only by subscription to Science] is by Dutch and British researchers at Max Planck Institute for Psycholinguistics, the Center for Language Studies, Radboud University, and the Leverhulme Centre for Human Evolutionary Studies, University of Cambridge, headed by Michael Dunn. They analysed over 125 terms with more strict criteria for homology than Ruhlen had (they used structural rather than cognate data to prevent bias1), from 16 Austronesian and 15 Papuan languages in the west Pacific and Papuan area.
This was a good choice, since this is both an area where the dates of settlement and migration are known fairly well, and is also a region of major linguistic complexity. And what they came up with was that one can only go back around 8000 years ± 2000 years.
That's not bad, though. 8-10 thousand years ago was just after the late Pleistocene; the period ended by the low point of the last ice age. The seas were as much as 320 feet lower, and many of the islands of Melanesia and Polynesia could be walked between.
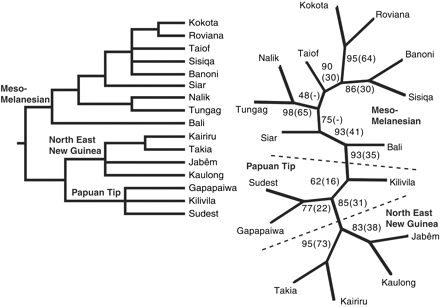 Dunn and colleagues found that the evolutionary tree of these languages suggested that the two major groups of the study - the Island Melanesian language group and the Papuan group - evolved from a common ancestor about that time, when the levels were lower. And an anomalous relationship of Bismark Islands languages to the rest can be explained by the fact that Bougainville and the Solomon Islands were united into a single landmass when the sea level was lower, while the Bismarks were always isolated by deep water.
The rates at which new words are coined and old words change turns out to be a power law distribution of sorts. This is what makes it hard to recover history past the depth of time Dunn and co. covered. It is also what makes it hard to work out long distant sequences of biological evolution, although genes and molecules chaneg far slower than languages. Russell Gray, in a commentary piece offers this graph:
Dunn and colleagues found that the evolutionary tree of these languages suggested that the two major groups of the study - the Island Melanesian language group and the Papuan group - evolved from a common ancestor about that time, when the levels were lower. And an anomalous relationship of Bismark Islands languages to the rest can be explained by the fact that Bougainville and the Solomon Islands were united into a single landmass when the sea level was lower, while the Bismarks were always isolated by deep water.
The rates at which new words are coined and old words change turns out to be a power law distribution of sorts. This is what makes it hard to recover history past the depth of time Dunn and co. covered. It is also what makes it hard to work out long distant sequences of biological evolution, although genes and molecules chaneg far slower than languages. Russell Gray, in a commentary piece offers this graph:
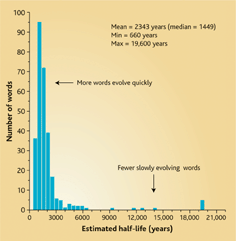 As time erases the homologies, history disappears. This means that there may be some things we will never know the course of. [That's no reason to think they didn't happen, though, even if we don't know them. Reality is not constrained by what we can know.]
People often think evolution is all about natural selection. But that is only one small part of it. The really interesting thing, at least to me, about evolution, is the evolutionary tree - phylogeny. And the methods used by biologists are general tools for understanding the past.
One commonly claimed disanalogy between cultural and biological evolution is that culture is horizontally as well as vertically transmitted. In biological terms, there is a lot of cross-lineage borrowing. But as a recent item noted, this is increasingly recongised in biology, too, and it is noteworthy that this does not seem to have muddied the phylogenetic "signal" in this case. History is lost, but some is retained. We need not become pure pessimists about knowing the past, in biology or culture.
1. Dunn M, Reesink G, Terrill A. 2002 "The East Papuan languages: A preliminary typological appraisal" Oceanic Linguistics 41 (1): 28-62 Jun - Thanks to Gary Hurd for the ref.
As time erases the homologies, history disappears. This means that there may be some things we will never know the course of. [That's no reason to think they didn't happen, though, even if we don't know them. Reality is not constrained by what we can know.]
People often think evolution is all about natural selection. But that is only one small part of it. The really interesting thing, at least to me, about evolution, is the evolutionary tree - phylogeny. And the methods used by biologists are general tools for understanding the past.
One commonly claimed disanalogy between cultural and biological evolution is that culture is horizontally as well as vertically transmitted. In biological terms, there is a lot of cross-lineage borrowing. But as a recent item noted, this is increasingly recongised in biology, too, and it is noteworthy that this does not seem to have muddied the phylogenetic "signal" in this case. History is lost, but some is retained. We need not become pure pessimists about knowing the past, in biology or culture.
1. Dunn M, Reesink G, Terrill A. 2002 "The East Papuan languages: A preliminary typological appraisal" Oceanic Linguistics 41 (1): 28-62 Jun - Thanks to Gary Hurd for the ref.
 Dunn and colleagues found that the evolutionary tree of these languages suggested that the two major groups of the study - the Island Melanesian language group and the Papuan group - evolved from a common ancestor about that time, when the levels were lower. And an anomalous relationship of Bismark Islands languages to the rest can be explained by the fact that Bougainville and the Solomon Islands were united into a single landmass when the sea level was lower, while the Bismarks were always isolated by deep water.
The rates at which new words are coined and old words change turns out to be a power law distribution of sorts. This is what makes it hard to recover history past the depth of time Dunn and co. covered. It is also what makes it hard to work out long distant sequences of biological evolution, although genes and molecules chaneg far slower than languages. Russell Gray, in a commentary piece offers this graph:
Dunn and colleagues found that the evolutionary tree of these languages suggested that the two major groups of the study - the Island Melanesian language group and the Papuan group - evolved from a common ancestor about that time, when the levels were lower. And an anomalous relationship of Bismark Islands languages to the rest can be explained by the fact that Bougainville and the Solomon Islands were united into a single landmass when the sea level was lower, while the Bismarks were always isolated by deep water.
The rates at which new words are coined and old words change turns out to be a power law distribution of sorts. This is what makes it hard to recover history past the depth of time Dunn and co. covered. It is also what makes it hard to work out long distant sequences of biological evolution, although genes and molecules chaneg far slower than languages. Russell Gray, in a commentary piece offers this graph:
 As time erases the homologies, history disappears. This means that there may be some things we will never know the course of. [That's no reason to think they didn't happen, though, even if we don't know them. Reality is not constrained by what we can know.]
People often think evolution is all about natural selection. But that is only one small part of it. The really interesting thing, at least to me, about evolution, is the evolutionary tree - phylogeny. And the methods used by biologists are general tools for understanding the past.
One commonly claimed disanalogy between cultural and biological evolution is that culture is horizontally as well as vertically transmitted. In biological terms, there is a lot of cross-lineage borrowing. But as a recent item noted, this is increasingly recongised in biology, too, and it is noteworthy that this does not seem to have muddied the phylogenetic "signal" in this case. History is lost, but some is retained. We need not become pure pessimists about knowing the past, in biology or culture.
1. Dunn M, Reesink G, Terrill A. 2002 "The East Papuan languages: A preliminary typological appraisal" Oceanic Linguistics 41 (1): 28-62 Jun - Thanks to Gary Hurd for the ref.
As time erases the homologies, history disappears. This means that there may be some things we will never know the course of. [That's no reason to think they didn't happen, though, even if we don't know them. Reality is not constrained by what we can know.]
People often think evolution is all about natural selection. But that is only one small part of it. The really interesting thing, at least to me, about evolution, is the evolutionary tree - phylogeny. And the methods used by biologists are general tools for understanding the past.
One commonly claimed disanalogy between cultural and biological evolution is that culture is horizontally as well as vertically transmitted. In biological terms, there is a lot of cross-lineage borrowing. But as a recent item noted, this is increasingly recongised in biology, too, and it is noteworthy that this does not seem to have muddied the phylogenetic "signal" in this case. History is lost, but some is retained. We need not become pure pessimists about knowing the past, in biology or culture.
1. Dunn M, Reesink G, Terrill A. 2002 "The East Papuan languages: A preliminary typological appraisal" Oceanic Linguistics 41 (1): 28-62 Jun - Thanks to Gary Hurd for the ref.
