3 recent reports use evolution to study mechanisms of antibody diversification
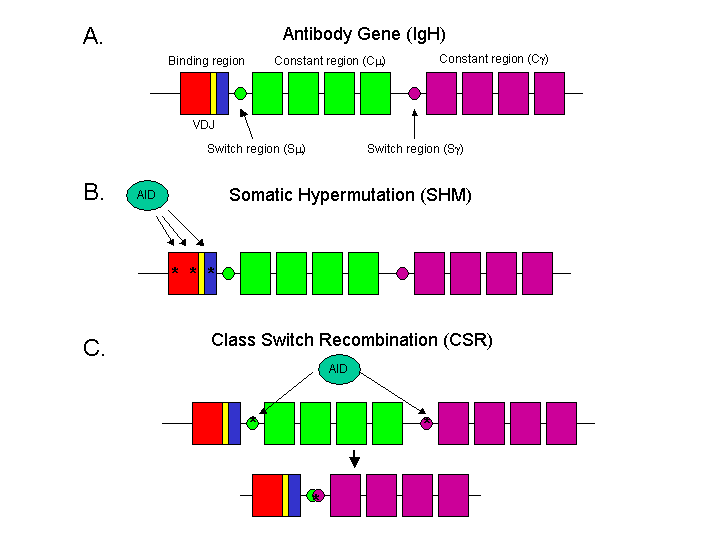
Figure 1 A. Genetic organization of a rearranged antibody heavy chain gene. The binding region, which is the rearranged VDJ exon is shown in red/yellow/blue. The 4 exons of the (mu) constant region are in green and the (gamma) constant region is in purple. The switch regions are represented by circles. B. In a putative model of somatic hypermutation, AID binds to the VDJ exon of the antibody and converts random cytosines to uracils (represented by asterisks). When the mismatch repair machinery repairs the mutated DNA, it utilizes an error-prone polymerase, often leaving a mutation near the site of conversion. C. In class switch recombination (CSR), AID targets the switch regions and converts cytosines to uracils there. The DNA repair machinery uses homologous recombination to repair the damage, causing the region between the two switch regions to be deleted, and changing the constant region exons.Evolution of CSR Like the V(D)J recombination system discussed here, it is not difficult to imagine how a three part irreducibly complex system like CSR could have evolved from another system. AID was already present in early tetropods, as SHM was and still is active in these organisms. The appearance of additional constant regions would only require a duplication of the original constant region, an entirely plausible event given that the bony fish that do not undergo CSR often contain multiple constant regions. In these organisms, the choice of which constant region is used is determined at the level of alterative splicing. Therefore, the only unique feature of the CSR system is the switch regions. Switch regions are extremely heterogenous in sequence, and have very little similarity to one another. The only feature they share in common is a propensity to form loop-like structures when in a single strand (such as when they're undergoing transcription). In fact, in some species of frog, switch regions are merely a region of repetitive AGCT tetramers, which don't even form loops. It isn't difficult to imagine a scenario by which AID could be co-opted from an SHM system for use in a primitive CSR system, swapping the original constant region with a duplicated one. But what about the switch regions? How will AID target the constant regions if no switch regions are present? The binding regions of the antibody genes don't contain sequences that resemble switch regions, yet they are ample targets for AID-mediated cytosine deamination, indicating that switch regions wouldn't even be necessary for a primitive CSR system. It's entirely possible that the switch regions evolved after CSR, providing better targets for AID. Because they don't have a research program of their own, IDists would be quick to play the incredulous card, doubting the feasibility of the scenario and demanding an unreasonable burden of proof to prevent them from dismissing the evolvability of the system altogether. While there are certainly gaps to be filled, no one denies that, the point of science is that the gaps can be filled. This is unlike ID, where no research is ever proposed and certainly never, ever conducted. What they did One issue with the evolution of CSR from SHM is whether the original AID in the early tetropods was capable of mediating a CSR-type reaction. The C-terminal region of AID is required for CSR, but not SHM, suggesting that additional components may be necessary to go from an SHM-capable AID protein to a CSR-capable one. In a recent issue of the Journal of Experimental Medicine, Vasco Barreto and colleagues from Michel Nussenzweig's lab reported that the AID gene from zebrafish was capable of activating CSR when transfected into mouse B cells that lacked their own AID gene. In this simple experiment, Barreto et al. demonstrated that even though bony fish do not possess a CSR system, their AID was fully able to catalyze CSR should the other components of the CSR system appear. They also examined pufferfish AID and found extremely low levels of CSR. In a similar study, published in the journal International Immunology, Wakae and collegues in the lab of Masamichi Muramatsu (who discovered AID while a post-doc in Tasuko Honjo's lab), tested both zebrafish AID as well as catfish AID, and found similar results. With these two studies, there could be no doubt that that particular step in the proposed evolution of the CSR system was entirely feasible. Additionally, it demonstrated that AID's catalytic mechanism for SHM was similar enough to CSR that it would be preserved in bony fish even after 400 million years of differential selection. Travis Ichikawa and colleagues, in the lab of PT's own Andrea Bottaro reported a more detailed analysis of pufferfish and frog AID in the Journal of Immunology. While frog AID was fully capable of mediating CSR in mouse AID-deficient B cells, they found essentially no CSR when pufferfish AID was transfected (remember, frogs have CSR, but pufferfish don't). Ichikawa et al. discovered that the catalytic domain of pufferfish AID was CSR competent, but the non-catalytic domain was inhibiting the reaction. As the C-terminal non-catalytic domain contains a nuclear export signal (NES) required for CSR activity (but not SHM) in mice, it was hypothesized that the pufferfish NES was somehow incompatible with mouse CSR. However, Ichikawa et al. disproved that notion by first observing that pufferfish AID had no difficulties in nuclear export/import in mouse B cells, and second that when replacing the NES sequence on human AID, the pufferfish NES did not negatively impact CSR. Why is this significant? While the details of class-switching and somatic hypermutation are of interest to only a handful of scientists, this research is significant to the ID/evolution debate in several ways. First of all, we now have clear-cut evidence of how a three part IC system could have evolved. No modifications would be necessary to convert an SHM-catalyzing AID gene into a CSR-catalyzing one. This provides yet another crystal clear example of co-option that can be widely cited for years to come. On another level, this research shows that more and more researchers are utilizing concepts in evolution to provide functional insight into their own areas of interest. The standard approach to characterizing a gene is to make mutations in important domains and see how that affects the function of the protein. However, by swapping pieces with another organism, this can identify important functional domains that no one might have thought to make mutations in. Furthermore, it helps to form a model for the evolution of the system, and this can lead to more functional predictions. Function informs evolution, and evolution informs function. Contrast this with ID. Because ID, as currently formulated by its chief advocates, places no constraints on the abilities or tendencies of the designer, all data is equally probable and equally consistent. Therefore, no functional predictions can be made from ID, and no functional data could ever provide evidence for or against ID. This is the primary reason why ID's most vociferous advocates don't do any research on ID. This brings up another reason for the significance of these reports. You may have heard Behe or Wells propose some experiments on ID. Aside from the fact that those experiments have nothing to do with ID, you've never heard either of them actually do the experiments they proposed. It's one thing to sit around and come up with ideas, it's another thing entirely to act on them. It costs nothing to propose an experiment, but to conduct one takes money, resources, and a few years of blood and sweat. That is the real B.S. detector for scientists. If their experiments are so great, why aren't they doing them? Here we have a terrific example of someone who posts regularly at the Panda's Thumb throwing his hat into the ring and doing an evolution-inspired experiment. Let's see Behe or Dembski or Wells do that. References 1. Barreto VM, Pan-Hammarstrom Q, Zhao Y, Hammarstrom L, Misulovin Z, Nussenzweig MC. AID from bony fish catalyzes class switch recombination. J Exp Med. 2005 Sep 19;202(6):733-8. 2. Wakae K, Magor BG, Saunders H, Nagaoka H, Kawamura A, Kinoshita K, Honjo T, Muramatsu M. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int Immunol. 2006 Jan;18(1):41-7. 3. Ichikawa HT, Sowden MP, Torelli AT, Bachl J, Huang P, Dance GS, Marr SH, Robert J, Wedekind JE, Smith HC, Bottaro A. Structural phylogenetic analysis of activation-induced deaminase function. J Immunol. 2006 Jul 1;177(1):355-61. Link to PDF
