Multiple codes in DNA
In an earlier posting, I promised to provide an overview of alternative codes in DNA. Such examples include alternative splicing and alternative reading frames (ARFs) which I will discuss here
The classical view of DNA was straightforward, DNA gets transcribed into RNA, introns get removed and the resulting exons form a protein. Exons are coding sequences in genes, introns are pieces of DNA/RNA that interrupt exons. The first step involves RNA synthesis (transcription) where the DNA is transcribed into RNA and exons and introns are still present, the next step is RNA splicing where introns are being removed. Alternative splicing causes different exons to be combined into messenger RNA (mRNA). These mRNA are translated into proteins in a step called protein synthesis.
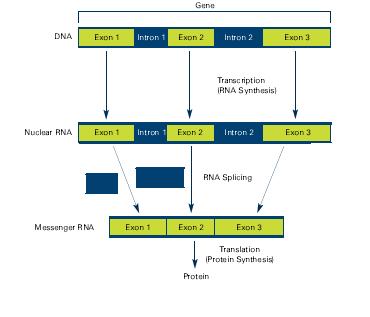
Alternative splicing involves different exons and result in different proteins from the same gene
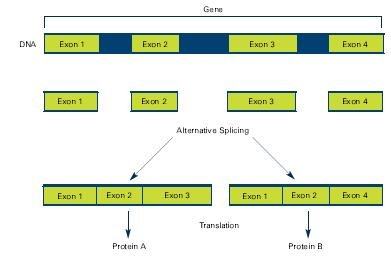
Source: The New Genetics NIH Publication No. 07-662
Alternative reading frames (ARFs) are overlapping stretches of DNA which encode for different proteins. Alternative reading frames are offset +1 or +2 and read in the same direction.
In general there are 6 reading frames
Once the RNA has been transcribed, it travels from the DNA template to the ribosome on the endoplasmic reticulum to be translated for protein synthesis. Each 3 bases in the RNA sequence codes for 1 amino acid. As you may not be sure what position to start at when predicting what protein sequence may be produced by this code, you could start with one of 3 positions from either end of the RNA sequence. Thus there are 6 possible predicted protein sequences resulting from such a piece of code. These are known as the 6 possible reading frames. There are 3 forward frames and 3 reverse sense frames.
Let’s perform an example
Assume you have just found a new stretch of DNA and you need to determine where to find an Open Reading Frame (ORF). An open reading frame starts with an atg (Met) in most species and ends with a stop codon (taa, tag or tga).
5’ TCAATGTAACGCGCTACCCGGAGCTCTGGGCCCAAATTTCATCCACT 3’
There are six possible interpretations, from right to left and left to right and with an offset of 0, 1 and 2.
Biology WorkBench is a tool which allows you to enter DNA sequences and find the ORF which has the longest frame.
Frame 1 start from left to right with zero offset
S M * R A T R S S G P K F H P
tca_atg_taacgcgctacccggagctctgggcccaaatttcatccact
Note the atg which is a start codon and taa which is a stop codon
Frame 2, offset 1
Q C N A L P G A L G P N F I H
caatgtaacgcgctacccggagctctgggcccaaatttcatccact
No start or stop codon
Frame 3, offset 2
N V T R Y P E L W A Q I S S T
aatgtaacgcgctacccggagctctgggcccaaatttcatccact 47
No start or stop codon
Frame 4, right to left zero offset
S G * N L G P E L R V A R Y I
agtggatgaaatttgggcccagagctccgggtagcgcgttacattga
One stop codon, no start codon
Frame 5, right to left one offset
V D E I W A Q S S G * R V T L
gtggatgaaatttgggcccagagctccgggtagcgcgttacattga
One stop codon, no start codon
Frame 6, right to left two offset
W M K F G P R A P G S A L H *
tgg_atg_aaatttgggcccagagctccgggtagcgcgttacattga
The sixth ORF is the longest one.
In “A First Look at ARFome: Dual-Coding Genes in Mammalian Genomes”, Chung et al, show that dual coding genes are not as rare in mammals as initially expected.
Coding of multiple proteins by overlapping reading frames is not a feature one would associate with eukaryotic genes. Indeed, codependency between codons of overlapping protein-coding regions imposes a unique set of evolutionary constraints, making it a costly arrangement. Yet in cases of tightly coexpressed interacting proteins, dual coding may be advantageous. Here we show that although dual coding is nearly impossible by chance, a number of human transcripts contain overlapping coding regions. Using newly developed statistical techniques, we identified 40 candidate genes with evolutionarily conserved overlapping coding regions. Because our approach is conservative, we expect mammals to possess more dual-coding genes. Our results emphasize that the skepticism surrounding eukaryotic dual coding is unwarranted: rather than being artifacts, overlapping reading frames are often hallmarks of fascinating biology.
Remember this paper which was quote-mined by Casey Luskin?
The paper shows the following figure (click to enlarge) which is Figure 2 in the original paper)
Notice how there is and ORF and an ARF, where ORF is the larger reading frame and ARF a subset, both ORF and ARF have a start and stop codon.
Addition
The ENCODE project proposes a new definition of gene
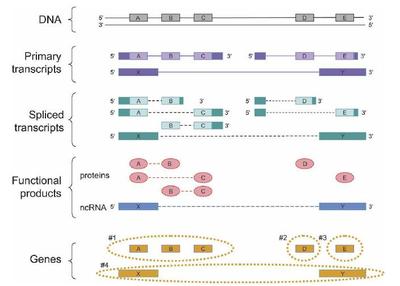
Figure 5. How the proposed definition of the gene can be applied to a sample case. A genomic region produces three primary transcripts. After alternative splicing, products of two of these encode five protein products, while the third encodes for a noncoding RNA (ncRNA) product. The protein products are encoded by three clusters of DNA sequence segments (A, B, and C; D; and E). In the case of the three-segment cluster (A, B, C), each DNA sequence segment is shared by at least two of the products. Two primary transcripts share a 5 untranslated region, but their translated regions D and E do not overlap. There is also one noncoding RNA product, and because its sequence is of RNA, not protein, the fact that it shares its genomic sequences (X and Y) with the protein-coding genomic segments A and E does not make it a co-product of these protein-coding genes. In summary, there are four genes in this region, and they are the sets of sequences shown inside the orange dashed lines: Gene 1 consists of the sequence segments A, B, and C; gene 2 consists of D; gene 3 of E; and gene 4 of X and Y. In the diagram, for clarity, the exonic and protein sequences A have been lined up vertically, so the dashed lines for the spliced transcripts and functional products indicate connectivity between the proteins sequences (ovals) and RNA sequences (boxes). (Solid boxes on transcripts) Untranslated sequences, (open boxes) translated sequences.
Mark B. Gerstein, Can Bruce, Joel S. Rozowsky, Deyou Zheng, Jiang Du, Jan O. Korbel, Olof Emanuelsson, Zhengdong D. Zhang, Sherman Weissman, and Michael Snyder What is a gene, post-ENCODE? History and updated definition Genome Res., Jun 2007; 17: 669 - 681.