An Open Letter to Dr. Michael Behe (Part 4)
Dear Dr. Behe
I’m sorry you couldn’t follow Ms Smith’s argument. I found it quite easy, an elegant detective story that built up its case clue by clue. However, even if you couldn’t follow it, the viroporin story was pretty hard to miss. That Vpu evolved over the space of a decade, when viral numbers were low, into a viroporin, a multisubunit structure with a function previously absent from HIV-1, was an obvious key challenge to your assertions.
I’m sorry that you didn’t understand the context of her evidence about Vpu variability. The fact that Vpu has undergone more sequence change than the envelop proteins, which have a high rate of evolution as they evade the bodies anti-vital defences, is a clue that they are evolving. What kind of evolution? This segued into her evidence about the function of Vpu, which arose recently in chimpanzees (see the diagram at the bottom of this post, which shows where Vpu arose), using this as a stepping stone to Vpu becoming a Viroporin in HIV-1 M.
Yes, you did state that you specifically excluded viral host-protein interactions (except when you can’t find any examples of such interactions, then, in the example of viral protein binding to host cell receptors, you claim the absence of new receptor binding supports your position, pity about the evolution of CXCR4 co-receptor binding then). I’m sorry, such handwaving doesn’t help you.
Firstly, Vpu viroproin is an example of viral protein-viral protein interaction. Secondly, as I show in the post to which you are supposed to be responding, the claim that viral protein –host protein interactions just “gum up the works” is nonsense. We are talking about a coordinated binding to a specific acceptor site (in what possible way is the development of binding of viral gp120 to CD4 in any way different to binding of class II MHC to CD4, they even use much the same binding residues (Wang 2002)). This is exactly the kind of binding site interaction that you claim can’t develop (or rather is incredibly improbable to develop).
Thirdly, your claims about the difficulty of developing binding sites rests almost entirely on the difficulty of getting multiple simultaneous mutations, the down stream effects of a new binding site are irrelevant to your argument . But even so, when we talk about Vpu binding we are talking about a coordinated binding to a specific acceptor site. In the context of the Vpu function, this is the production of a multi-subunit molecular machine that sends CD4 to be broken down. The recently evolved Vpu binding sites change the targeting of this complex, with beneficial effects for the virus.
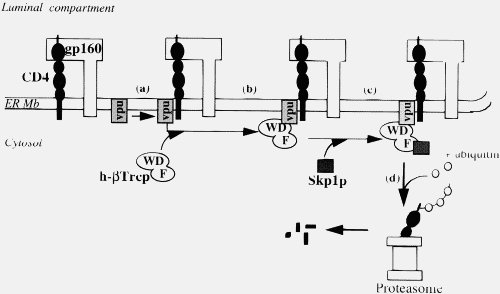
Vpu, no, it doesn’t “gum up the works”
You use a similar non-sequitur when it comes to the development of the casein kinase binding sites. You claim they are too simple, but your own example of a binding site is the haemoglobin S mutation, a single binding site. Your book categorically states no new binding sites in HIV, contrasting this with the one HbS one amino acid binding site mutation. At the very least, in the interests of honesty, in your book you should have noted the casein kinase binding sites and stated they were “too simple” for you (even though they require more than a single point mutation, which is where you claim things ramp up in difficulty, and they are more complex than your HbS example).
The casein kinase data should give you pause anyway. These are key, regulatory protein-protein binding sites. Activation and regulation of diverse proteins are dependent on these regulatory binding sites, and you claim they can develop easily. Yet they are no different to any other protein-protein binding site, and have a quite nice affinity for their enzyme. Might not the casein kinase site be a paradigm of how protein-protein binding develops, via multiple paths and through selectable intermediates? I would suggest reading Wang (2002) again, and paying particular attention to the role of amino acid F43 in binding to CD4.
Still, you admit you did see the viroporin section. This is a direct challenge to the very heart of your argument. Why did you ignore it?
Yours sincerely
A male featherless biped named Ian Musgrave
References:
Wang J. (2002) Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem Sci. Mar;27(3):122-6.
