The Revenge of the Guinea Pigs
The nested hierarchy of DNA/Protein sequence similarity is powerful evidence for common descent. The parallel hierarchy formed by broken genes is more powerful still. Even anti-evolutionist Dr. Michael Behe accepts it as evidence of common descent. Behe’s acceptance of the evidence from broken genes confuses the conventional creationists, as seen in a report of a recent creationist forum. However, the creationists think they have a way out, but wouldn’t you know it, they are wrong.
Creatures inherit their DNA from their parents, who have inherited their DNA from their parents and so on into the past. We use this information to do paternity tests, and form genealogies. The nested hierarchy of DNA similarity is evidence that organisms descended from a common ancestor. Creationists try to deny this of course, and often invoke the idea that the similarity between organisms proteins reflects common design, that the proteins are similar because they have similar function. They never explain why, for example, the DNA sequences of the Tasmanian Tiger, a carnivorous marsupial that fills the niche of wolves (and looks pretty wolf-like), is much closer to that of the vegetarian Kangaroo than to the wolves, dogs and foxes whose ecological role and function they duplicate. Or why housekeeping genes with the same function, which are not dependent on the animals environment, also from this hierarchy.
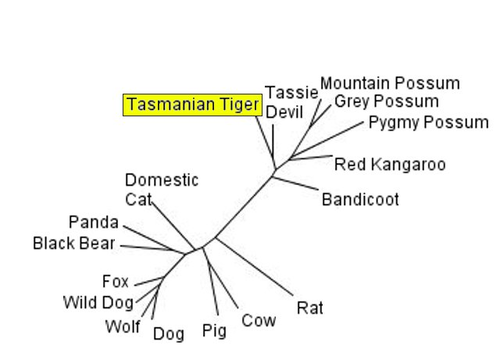 Phylogeny based on the sequence cytochrome b. The mid-sized carnivore the Tasmanian Tiger is shown to be more closely related to the vegetarian Kangaroo, and bandicoots and possums, than to its ecological niche and functional equivalents the dog/wolf family. As well, the sequence of the vegetarian Panda is more similar to carnivorous Bears than to other herbivorous animals
Phylogeny based on the sequence cytochrome b. The mid-sized carnivore the Tasmanian Tiger is shown to be more closely related to the vegetarian Kangaroo, and bandicoots and possums, than to its ecological niche and functional equivalents the dog/wolf family. As well, the sequence of the vegetarian Panda is more similar to carnivorous Bears than to other herbivorous animals
Despite the glaring flaws in the creationist “similar function” argument, it would good to have another line of evidence for ancestry. This evidence comes from broken genes (pseudogenes) in our genome. The genome is littered with non-functional sequences (indeed there are more non-functional sequences than functional genes), ranging from parasitic non-coding sequences like LINES to old broken viruses and broken genes. Over time, these sequences acquire mutations, organisms that share a recent common ancestor will have accumulated fewer mutations than organisms that have a distant common ancestor. These sequences also form a nested hierarchy that parallels the pattern we see with functional genes. It is very hard to argue that a nested hierarchy of broken genes represents common design (unless the Designer is the Sirius Cybernetics Corporation). The classical example of a broken gene in the human lineage is the gene L-gulono-gamma-lactone oxidase (GULO), which catalyses the last step of ascorbic acid biosynthesis. It is broken in primates, hence we get scurvy when we don’t eat fresh fruit or vegetables. Aligning the sequences of primate GULO genes shows humans being related to primates, with the chimpanzee being our closest relative.
As I mentioned, the nested hierarchy of the broken version of the GULO gene is powerful evidence of common ancestry. The Guinea Pig also has a broken GLUO gene, but it is broken in a different way to the primate genes, and does not nest with them. Dr. Michael Behe accepts data such as the nested hierarchy of the broken GULO genes as evidence of common descent. This confuses the creationists (isn’t Behe on our side?). At a recent creationist forum Paul Nelson tried to help a such a confused person by attempting to explain away the GULO evidence. However, his “explanation” was based on an error.
In 2003 the Nishikimi group compared the sequences of primate, Guinea Pig and rat GULO genes. The found that some sections of the Guinea Pig sequence matched the primate sequences more closely than rat sequences. They proposed that there were “mutational hotspots” in the GULO allowing convergent evolution between Guinea Pig and primate sequences. Some creationists immediately jumped on this paper, claiming that these alleged “mutational hotspots” invalidated common descent. This is a complete furphy. As mentioned above and as can be seen from the phylogenic tree below, the Guinea Pig nests more closely with rodents, and nowhere near the primates.
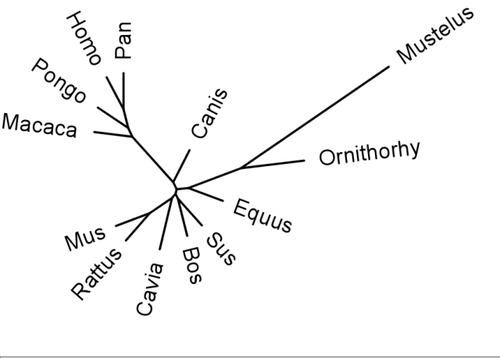 Phylogenic tree of GULO sequences. As can be seen, the primate sequences nest together, with the chimpanzees and humans closest, as has been found with other genes. Guinea Pigs (Cavia) are closer to rodents (Mus; mouse and Rattus; Rat) than primates. Sequences aligned with MAFF and trees generated with PHYLIP.
Phylogenic tree of GULO sequences. As can be seen, the primate sequences nest together, with the chimpanzees and humans closest, as has been found with other genes. Guinea Pigs (Cavia) are closer to rodents (Mus; mouse and Rattus; Rat) than primates. Sequences aligned with MAFF and trees generated with PHYLIP.
One problem with the “mutational hotspot’ idea is that it is completely wrong. To claim that Guinea Pig and primate sequences have converged, you have to compare them with a sequence that is more representative of the ancestral sequence. Nishikimi’s group compared Guinea Pig and primate to rat sequences. However, it is the rat that has evolved significantly from the ancestral sequence, not the Guinea Pig and primate that have converged.
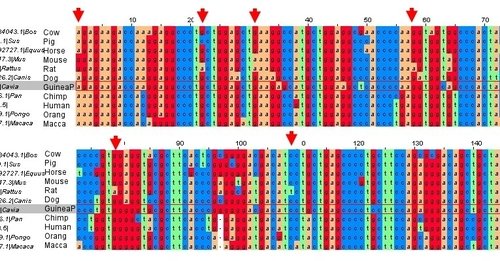 Alignment of mammalian GULO sequences (click to get larger version). The arrows indicate nucleotide positions where Nishikimi (2003) claimed that Guinea Pig and primate sequences had converged. As can be clearly seen, it is Guinea Pig (highlighted) and primate that retain the ancestral sequences, and the rat has diverged from this common sequence.
Alignment of mammalian GULO sequences (click to get larger version). The arrows indicate nucleotide positions where Nishikimi (2003) claimed that Guinea Pig and primate sequences had converged. As can be clearly seen, it is Guinea Pig (highlighted) and primate that retain the ancestral sequences, and the rat has diverged from this common sequence.
As can be clearly seen in the above diagram, the rat sequence has diverged from the ancestral sequences. So no mutational hotspots; Nishikimi was incorrect (and has admitted so in private correspondence). But that’s the good thing about science, it is self correcting, if we make mistakes, we fix them when new data comes out.
What gets me is that creationists could have done this analysis of GULO genes themselves (like the other analysis I have reported on). I used entirely public databases and web-based programs to do the trees and alignments. The Creationists did nothing, and are propagating an error.
