Plaque--evidence for Design!
Every now and then, I check in over at The Institute for Genomic Research (TIGR) to see what new projects they’re up to, as well as to see if they’ve released a particular genome sequence I’m waiting on. Yesterday I noticed this project:
Innovative Metagenomics Strategy Used To Study Oral Microbes
Rockville, MD - The mouth is awash in microbes, but scientists so far have merely scratched the surface in identifying and studying the hundreds of bacteria that live in biofilm communities that stick to the teeth and gums.
In an innovative new project that could help improve the detection and treatment of oral diseases, scientists are now using a metagenomics strategy to analyze the complex and difficult-to-study community of microbes in the oral cavity.
***
In recent years, molecular methods have indicated that there are well over 400 species of bacteria in the oral cavity. But, so far, only about 150 of those species have been cultured in laboratories and given scientific names. Using a metagenomics sequencing strategy, TIGR scientists will be able to identify bits and pieces of the DNA of many of those oral microbes that so far have not been grown in labs and studied.
Now, I know that there are an insane amount of microbes in the mouth, but 400 species? Holy cow.
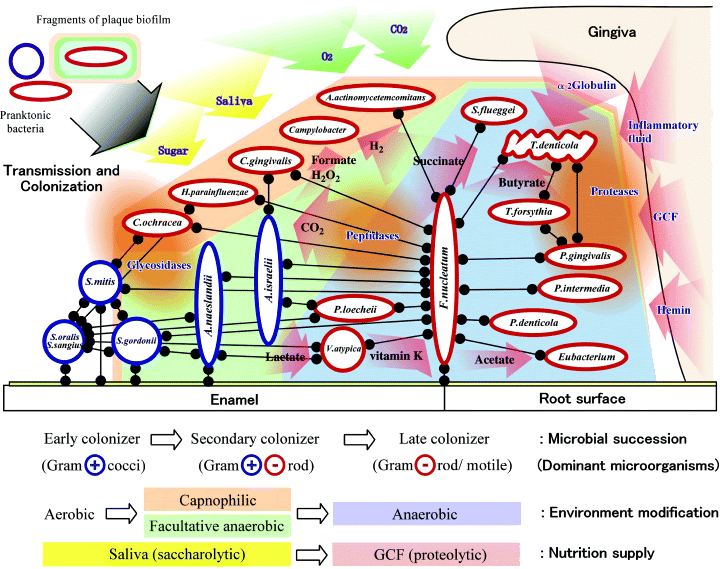
For anyone who may be unfamiliar with the microbiology of plaque, it’s an incredibly complex mixture consisting of both bacterial and host cells, as well as inorganic material such as calcium and phosphate. The formation of plaque has been fairly well-studied, and can be broken down into several steps.
1) Pellicle formation. This begins as soon as teeth are cleaned. Substances (mainly glycoproteins) in your saliva then adhere to your teeth.
2) In the next step, the pellicle-coated teeth is colonized by a number of gram-positive bacteria, including several strains of Streptococci. These “primary colonizers” produce proteins which allow for attachment and adherence to the pellicle-coated tooth surface.
3) Plaque on the surface then increases as the primary colonizers divide, and as secondary colonizers attach to the primary colonizers, and multiply in number as well. Secondary colonizers include gram-negatives such as Prevotella intermedia and Capnocytophaga species (as well as many others).
4) Tertiary colonizers (generally ~1 week post-brushing–so hopefully most of us aren’t too familiar with them!) These include species of Porphyromonas, Campylobacter, and Treponema, and again, many others. By this time, metabolic reactions in the biofilm may lead to predictable structures in some members of the plaque: for example, a “corn cob” like array of cocci bound to filaments within the biofilm.
While some of the bacteria present feed off of oral nutrients supplied by the host (such as sugars), others within the plaque rely upon nutrients produced by other bacteria within the biofilm. Additionally, some species produce products which inhibit the growth of other species in the biofilm; for example, peroxide or acids. And of course, these acids also play a role in producing the characteristic holes in your teeth that can come from regular accumulation of dental plaque.
Nishihara and Koseki provide this schematic of the current model of dental plaque formation:
Of course, plaque are not the only biofilm we know of. They’re everywhere: on your shower curtain, on medical devices implanted in patients, on rocks in rivers and streams, and in your nose. While the sheer number of different organisms a biofilm may contain makes it a challenge to study, I personally relish that challenge, and would much rather tackle it than throw my hands in the air and say it’s “too complex” to have come about naturally.

