Celebrating Darwin Day by Correcting Michael Behe on the Evolution of Polar Bears
This guest post by Nathan Lents is cross-posted from The Human Evolution Blog. It was co-authored with Arthur Hunt of the University of Kentucky, who first pointed out some of these errors on the Peaceful Science forum. I wish I had spotted these myself before Richard Lenski, Josh Swamidass, and I wrote our review for Science, but I took Behe's word on the polar bears because it all sounded solid. In other words, I did what Behe hopes all his readers will do - just believe him and not check the reference. Lesson learned, and kudos to Art for catching this and for working with me on this post. Matt Young is the moderator of the post on PT. We have discussed the Science review here
The release of Michael J. Behe’s newest book, Darwin Devolves: The New Science About DNA That Challenges Evolution, is nearly upon us and so the first chapter was made publicly available to entice readers. In this chapter, Behe outlines his main thesis: at the molecular level, adaptive changes are largely due to events that in some way destroy or damage proteins and enzymes. He calls it the first rule of adaptive evolution and to illustrate his point, he discusses the evolution of polar bears and describes the molecular events in that evolution as nothing more than a series of damaging mutations that result in a more adapted organism.

But first, a quick introduction to Behe for those who may not know who Behe is or where this is coming from. With the release of his first book, Darwin’s Black Box, in 1996, Behe helped revolutionize and reorganize the resistance to modern evolution under the banner known as “Intelligent Design,” often abbreviated as ID. Many consider ID as simply creationism by another name, but the ID community works hard to distance themselves from that label. They insist that ID is a scientific theory, not a religious one, based on what they consider evidence that cells and organisms were designed intentionally, rather than the result of the aimless and unguided forces of evolution. Scientists and federal courts disagree, but this has not stopped the steamroller of pseudoscientific claims from the ID community.
Fast forward to 2019 and Behe is at it again. In Darwin Devolves, Behe makes the argument that natural selection, which he prefers to call “Darwinism,” is driven largely, even exclusively, by mutations that degrade or destroy protein function. At the outset, it must be said that we have known for many decades that, occasionally, harming or even destroying a gene or protein can actually be good for the organism. What Behe is saying is that harming genes is the only way that unguided mutations can ever help an organism. That’s just not the case, but I’ll get to that later.

Back to the polar bears. Behe offers them as an example of how harming genes can help an organism and lead to adaptive evolution. Imagine an ancestor bear population that looked pretty much like brown bears. Then came some random mutations that reduced the production or deposition of pigment into the fur of the bears. This made the bears white and - voilà! - the bears acquired natural camouflage in snowy climates so as to better sneak up on their prey.
This seems like a pretty straightforward example and most people will simply take it at face value. Behe jumps from this example to his claim that this is all that unguided mutations can do. However, even in this apparently “pro-Darwinism” example, Behe exaggerates his claims and misrepresents what science has actually revealed. The evolution of polar bears was not only a matter of harmful mutations.

The key reference here is a 2014 paper in Cell. In this study, researchers did genome sequencing of 89 polar bears and brown bears and discovered the molecular changes that distinguish these very closely related species, using the giant panda as the reference sample. The results were fascinating. For example, the analysis revealed that polar bears and brown bears have been separate populations with limited gene flow for less than 500,000 years.
In the polar bears, the researchers found over a dozen protein-coding genes that had experienced recent positive selection, meaning that evolutionary forces had strongly favored specific variants. Brown bears, on the other hand, have experienced much less positive selection since the populations diverged. This means that polar bears have experienced stronger selection and have diverged from the ancestors more than brown bears have. This matches what the fossil evidence suggests. Basically, a population of brown bears ventured northward and, in response to the very different climate, evolved into polar bears. The ancestral population stayed pretty much the same and are the brown bears of today.
One of the genes that experienced the strongest selection is APOB, which encodes apolipoprotein B, a protein involved in the transport of fat molecules in the blood. This makes sense because polar bears subsist on a diet extremely rich in saturated fats, yet don’t develop heart disease with great frequency. Quoting Behe:
The polar bear’s most strongly-selected mutations – and thus most important for its survival – occurred in a gene dubbed APOB, which is involved in fat metabolism in mammals, including humans. That itself is not surprising, since the diet of polar bears contains a very large proportion of fat (much higher than in the diet of brown bears) from seal blubber, so we might expect metabolic changes were needed to accommodate it.
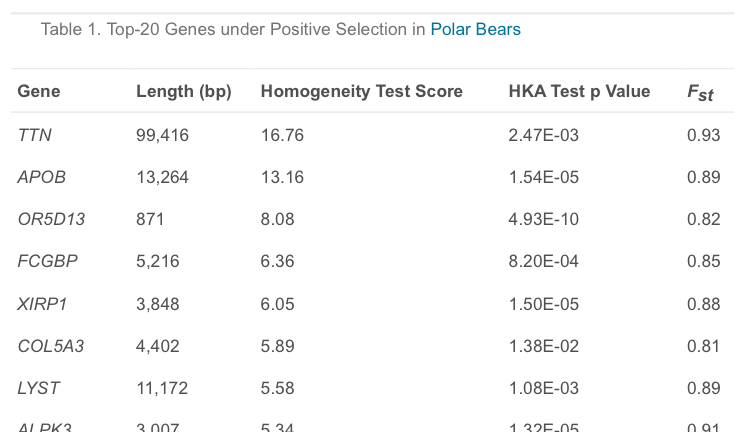
First of all, as shown in Table 1 of the paper, APOB harbors the second most strongly-selected set of variants, not the first, but we can let that one slide.
Behe continues:
But what precisely did the changes in polar bear APOB do to it compared to that of other mammals? When the same gene is mutated in humans and mice, studies show it frequently leads to high levels of cholesterol and heart disease. The scientists who studied the polar bear’s genome detected multiple mutations in APOB. Since few experiments can be done with grumpy polar bears, they analyzed the changes by computer. They determined that the mutations were very likely to be damaging -that is, likely to degrade or destroy the function of the protein the gene codes for.
Some of them, yes. Definitely not all of them or even most of them. He continues:
A second highly-selected gene, LYST, is associated with pigmentation, and changes in it are probably responsible for the blanching of the ancestors’ brown fur. Computer analysis of the multiple mutations of the gene showed that they too were almost certainly damaging to its function. In fact, of all the mutations in the seventeen genes that were most selected, about half were predicted to damage the function of the coded proteins. Furthermore, since most altered genes bore several mutations, only three to six (depending on the method of estimation) out of seventeen genes were free of degrading changes. Put differently, 65-83 percent of helpful, positively selected genes are estimated to have suffered at least one damaging mutation.
Now it’s getting harder to excuse Behe’s exaggeration. Specifically, the authors found that only 7 of the 17 genes with the strongest signatures for positive selection are unequivocally predicted to possess at least one “damaging” mutation. Even Behe’s “about half” is just 41%, which means that the lower limit on Behe’s estimation is also wrong. It’s not 65-83%, it’s 41-83%. The range is so wide because computational predictions invariably involve uncertainty.
But more importantly, by Behe’s math, if a gene harbors five enhancing mutations and one diminishing one (by computer prediction), it counts as “damaged.” Any number of gain-of-function mutations can be overtaken by a single damaging one. While that could be true in some cases, a nonsense mutation for example, there is no reason to assume it must be true in all cases. (Unless, of course, you have a specific argument you’re trying to make.)
Considering how little is known about the molecular biology of polar bears, it is entirely possible that none of the 17 most positively selected genes in polar bears are “damaged.” Quoting from the Supplemental information in the paper:
We find no fixed missense mutations specific to the polar bear lineage associated with human diseases according in the Human Gene Mutation Database. However, the top 20 genes are significantly enriched with genes previously associated with metabolic diseases and traits and humans (p-value = 0.042) from the GWAS catalog, discussed in the main text.
In other words, many of the 20 most positively selected genes in the polar bear genome are orthologs of genes that have variants (mutations) in the human population associated with metabolic disease. So when these genes are damaged by mutations in humans, the humans are more likely to suffer metabolic diseases. But Behe believes - with no hard evidence mind you! - that these same genes, when damaged, protect the polar bears from metabolic disease.
And there’s more. If we come back to APOB, the polar bear gene that Behe spends the most time discussing, we find that the authors of the study have a very different interpretation of the data than Behe does. Quoting the paper again [emphasis added]:
Substantial work has been done on the functional significance of APOB mutations in other mammals. In humans and mice, genetic APOB variants associated with increased levels of apoB are also associated with unusually high plasma concentrations of cholesterol and LDL, which in turn contribute to hypercholesterolemia and heart disease in humans (Benn, 2009; Hegele, 2009). In contrast with brown bear, which has no fixed APOB mutations compared to the giant panda genome, we find nine fixed missense mutations in the polar bear (Figure 5A). Five of the nine cluster within the N-terminal ba1 domain of the APOB gene, although the region comprises only 22% of the protein (binomial test p value = 0.029). This domain encodes the surface region and contains the majority of functional domains for lipid transport. We suggest that the shift to a diet consisting predominantly of fatty acids in polar bears induced adaptive changes in APOB, which enabled the species to cope with high fatty acid intake by contributing to the effective clearance of cholesterol from the blood.
Clearly, the authors do not expect the polar bear APOB to be “broken.” Rather, a bare majority of the amino acid changes are in the most important region for the clearing of cholesterol from the blood. In other words, these mutations likely enhance the function of apoB, at least when it comes to surviving on a diet high in saturated fats.
It is also worth noting that apoB does much more than clear fatty acids from the blood. It is a very large protein that has many biochemical activities and is a central player for lipid and cholesterol transport. Even if “damaging” mutations might be beneficial in one context, they could very well be harmful or lethal in another. Moreover, mice that lack apoB are not viable.
To recap: 1.) There is no evidence for Behe’s claim that APOB is degraded or diminished in polar bears and everything we know about the protein from other mammals suggests the opposite. And 2.) Behe’s claim that the most common adaptive changes in polar bears are those that degrade or destroy proteins is not supported, and the evidence suggests otherwise. Those are just the errors that we found in his first example.
And yet Behe makes this bold claim:
It seems, then, that the magnificent Ursus maritimus has adjusted to its harsh environment mainly by degrading genes its ancestors already possessed. Despite its impressive abilities, rather than evolving, it has adapted predominantly by devolving. What that portends for our conception of evolution is the principal topic of this book.
If you are wondering what the word “devolving” means, join the club. I’ll leave that discussion for another day. (And that day will come soon…. stay tuned.)
For more of the problems with Darwin Devolves, see my review in Science (co-authored with Richard Lenski and Josh Swamidass, open-access version), my essay in Skeptic magazine (available in a couple weeks), and my book review in AiPT!